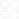
1:TheuseofrecombinantDNAtechnologybegansoonafterthediscoveryofDNAligaseandrestrictionendonucleases.Soonafter,adventofthepolymerasechainreaction(PCR)openedupnewpossibilitiesforamplificationofspecificDNAsequencesfromacomplexmixtureofgenomicDNA.ThesetechnologieshavebeenamainstayinthemodernscientificlaboratoryforseveraldecadesandremainusefulmethodsforcloningpotentiallyvaluableorinterestingDNAtoday.However,asscientistsseektoworkwithlargerDNAfragments,conductextensivere-engineeringofgeneticelements,synthesizewholegenomesandmovetowardsautomatedap-proaches,thetechnologiesrequiredtomanipulateDNAalsoneedtoevolve.InvestigatorsattheJ.CraigVenterInstitute(JCVI)havedevelopedanumberofinvitroenzymaticstrategiestoassembleshortoligonu-cleotidesintolargerdouble-strandedDNAcon-structs(1-4).In2003,JCVImadeasignificantadvancementintheproductionofasyntheticgenomebyassemblingthe5,386bpgenomeofphiX174,avirusthatinfectsbacteria,injust14days(5).Thisapproachinvolvedjoiningsyntheticoligonucleotidesbypolymerasecyclingassembly,andsubsequentlyamplifyingthembyPCR(5-6).Theunprecedentedspeedwithwhichthiswascompletedlaidthefoundationforcon-structinglargerandmorecomplexgenomes.In2004,JCVIbegansynthesizingtheMycoplas-magenitaliumgenome.Itwasfoundthatoverlap-pingDNAmoleculescouldbeefficientlyjoinedusingthreeenzymespecificities:(i)exonucleaseactivity,thatchewsbacktheendsofDNAfrag-mentsandexposesssDNAoverhangsthatcanannealtotheirssDNAcomplement;(ii)DNApolymeraseactivity,thatfillsgapsintheannealedproducts,and(iii)DNAligaseactivity,thatcova-lentlysealstheresultingnicksintheassembly.AGibsonAssemblyhasbecomethemostcommonlyusedoftheinvitroassemblymethodsdiscussedabove,asitiseasy-to-use,flexibleandneedslittleornooptimization,evenforlarge,complexassemblies.AllthatisrequiredisinputDNAwithappropriateoverlaps,andanappropriatemixofthethreeenzymes–theGibsonAssemblyMasterMix.DNAfragmentsareaddedtothemastermixandincubatedat50°Cfor1hour;theresult-ingassemblyproductisafullysealeddsDNAsuitableforarangeofdownstreamapplications(Figure1).JCVIhasusedGibsonAssemblytorapidlysynthesizetheentire16,520bpmousemitochondrialgenomefrom600overlapping60-baseoligonucleotides(3).Itwasalsousedincombinationwithyeastassemblytosynthesizethe1.1MbpMycoplasmamycoidesgenome,whichwasthenactivatedinarecipientcelltoproducethefirstsyntheticcell(1).GibsonAssembly™–BuildingaSyntheticBiologyToolsetInthequesttocreatethefirstbacterialcellcontrolledbyasyntheticgenome,theJ.CraigVenterInstitute(JCVI),withsupportfromSyntheticGenomics,Inc.(SGI),developedavarietyofpowerfulnewDNAsynthesisandassemblymethodologies(1–5)tomanipulatelarge,complexDNAs.Thesemethodsincludeasimple,one-stepisothermalinvitrorecombinationtechnologycapableofjoiningDNAsrangingfromrelativelyshortoligonucleotidestofragmentshundredsofkilobasesinlength.Thisapproach,commonlyreferredtoas“GibsonAssembly,”isnowbeingusedinlaboratoriesaroundtheworldtoconstructDNAfragments.Ithasthepotentialtoimproveupontraditionalcloningmethodsandopensuparangeofinnovativeandultimatelyveryusefulreal-worldapplications.FeatureArticletwo-stepthermocycle-basedinvitrorecombina-tionmethodutilizingtheseenzymeswasusedtojoin101overlappingDNAcassettesintofourpartsoftheM.genitaliumgenome,eachbetween136kband166kbinsize.Thismilestonemarkedthefirstassemblyofagenomederivedfromafree-livingorganism.At582,970bp,thissyntheticgenomewasthelargestchemicallyde-finedDNAstructuresynthesizedinalaboratory,andwas18timeslargerthananyDNAthathadpreviouslybeensynthesized(4).Sincethen,twoadditionalinvitrorecombinationmethodshavebeendevelopedbyJCVItojoinandcloneDNAmoleculeslargerthan300kbinasinglestep(2-4).ThesimplestofthesemethodsisGibsonAssembly,aone-stepisothermalap-proachthatutilizesthesamethreeenzymaticactivitiesdescribedpreviously.ThismethodcanbeusedtojoinbothssDNAanddsDNAs.Figure1.OverviewofGibsonAssembly.AAddfragmentstoGibsonAssemblyMasterMix.dsDNAfragmentswithoverlappingends.5´3´5´Exonucleasechewsback5´ends.3´5´3´5´5´3´DNAfragmentsanneal.DNApolymeraseextends3´ends.DNAligasesealsnicks.3´5´5´3´3´5´5´3´BFullyAssembledDNAA+BIncubateat50°Cfor1hour.GibsonAssembly2ApplicationsofGibsonAssembly:Cloning.GibsonAssemblyeliminatestheneedtoengineerrestrictionenzymecutsiteswithinDNAwhenassemblingfragmentstogether.DNAmol-eculesaredesignedsuchthatneighboringfrag-mentscontaina20-40bpoverlappingsequence.IftheDNAfragmentsoriginatefromPCRproducts,theoverlappingsequenceisintroducedatthe5′endsoftheprimersusedintheamplificationreaction(Figure2).DNAfragmentscanalsobeas-sembledwithrestrictionenzymedigestedorPCRamplifiedvectortoformcircularproductssuitableforcloning,orforuseindownstreamapplications,suchasrollingcircleamplification(RCA).ToproducethesevectorsbyPCR,eachprimerneedstoincludeanoverlapwithoneendofthevector,arestrictionsite(e.g.,NotI)notpresentwithintheinsertorinsertstoenableittobereleasedfromthevector,andanoverlapwiththeendsoftheDNAfragmentassemblyorinsert.JCVIhasbeenusingthisapproacht